Author : Dennis Núñez, Lamberto Ballan, Gabriel Jiménez-Avalos, Jorge Coronel, Mirko Zimic
July 2020 – https://www.researchgate.net/publication/342732879_Using_Capsule_Neural_Network_to_predict_Tuberculosis_in_lens-free_microscopic_images
Abstract
Tuberculosis, caused by a bacteria called My-cobacterium tuberculosis, is one of the most seri-ous public health problems worldwide. This workseeks to facilitate and automate the prediction oftuberculosis by the MODS method and using lens-free microscopy, which is easy to use by untrainedpersonnel. We employ the CapsNet architecturein our collected dataset and show that it has a bet-ter accuracy than traditional CNN architectures.
1. Introduction
Tuberculosis (TB), caused by the Mycobacterium tuber-culosis bacteria, is one of the most serious public health problems in Peru and worldwide. Around a quarter of the world’s population, are infected with TB and 1.2 million people died from TB in 2018 (WHO,2019).The MODS method, (Microscopic Observation Drug Sus-ceptibility Assay) (Rodr´ıguez et al.,2014) developed inPeru, allows the quick recognition of morphological pat-terns of mycobacteria in a liquid medium (directly froma sputum sample). It is a cost-effective option, since TBcan be detected in just 7 to 21 days, which is a fast and alow-cost alternative test. This method is included in the listof rapid tests authorized by the National Tuberculosis Pre-vention and Control Strategy. In addition, complementingthis method with lens-less microscopy is very important dueit is easy to use and to calibrate by no trained personnel.In recent years Convolutional Neural Networks (CNNs), aclass of deep neural networks, have become the state-of-the-art for object recognition in computer vision (Krizhevskyet al.,2012), and have potential in object detection (Arelet al.,2010) and segmentation (Krizhevsky et al.,2012)
For the analysis of medical images via deep learning, image classification is often essential. Manual evaluation by experienced clinicians, is important; however, it is laborious and time-consuming, and may be subjective. Anotherapproach for image classification, a fully convolutional neural network known as CapsNet (Sabour et al.,2017), hasrecently shown promising results (Kwabena Patrick et al.,2019). The CapsNet has been applied for some medical andmicroscopy images, including classification of 2D HeLa cells in fluorescence microscopy, classification of Apoptosisin phase contrast microscopy, and classification of breasttissue (Zhang & Zhao,2019;Mobiny et al.,2020;Iesmantas& Alzbutas,2018).The purpose of this work is to evaluate the feasibility ofautomatic TB detection on lens-free images. We applied theCapsNet architecture for image classification.
2. Methodology
For prediction of TB on grayscale images, the proposed methodology makes use of image patching followed by the CapsNet classifier in order to have an efficient classifier us-ing few training images. We have the following steps: Firstthe full image (3840×2700 pixels) is divided into sub-images(256×256 pixels), then each sub-image is passed forwardthe Caps Net classifier and provides the positive/negative label for each sub-image. Then, the histogram of classes forevery full image is constructed. Finally, given a full image, the logistic regressor makes the prediction of TB
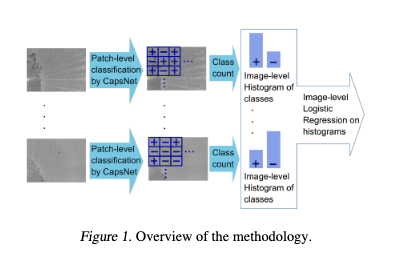

The images were collected by the iPRASENSE Cytonote lens-free microscope. These images were reconstructed from the holograms that were obtained by the microscope via the iPRASENSE software. As shown in Figure 2, the collected images have a dimension of 3840×2700 pixels and are presented in grayscale format. After data collection and manual annotation, we obtained 10 full annotated images.To augment this dataset and due the images are very large compared to the TB cords, we performed the division of the images into sub-images. So, each image was divided into sub-images of 256×256 pixels. Since some TB cords could be near the margin of each sub-image, we considered an overlapping of 20 pixels between adjacent sub-images.With the above mentioned considerations and taken the same number of images for both classes (areas without Tb cords in the full images are larger than the areas with TB cords),we obtained a total of 500 positive and 500 negative sub-images. These positive and negative images were used to train/test the different models, see Figure 3.
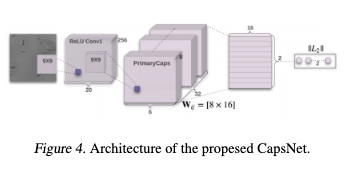
The annotation of the collected images was as follows: Firsta medical expert, which has extensive experience in TB,visually inspected each sub-image. If the medical expertfound at least one TB cord the sub-image was annotated aspositive, otherwise annotated as negative.The network is based on the CapsNet convolutional network,see Figure 4. There, the first two layers of the convolutionkernel we used for the training model are 9*9. The selectionof the convolution kernel is related to whether the low-level features can be accurately obtained. If the size of theconvolution kernel is too small, the selected features willbe enhanced accordingly, but if there is noise in the image,the noise will be enhanced, thus overwhelming the usefuldetails in the image.In addition, the Capsule Network performance is relativelystrong. On small datasets, CapsNet is significantly betterthan CNN. For very few samples, the capsule network stillhas good accuracy and convergence (Sabour et al.,2017)

3. Results and Discussion
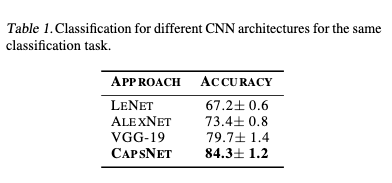
The CapsNet was trained over 450 positive and 450 nega-tive sub-images, and tested on 50 positive and 50 negative sub-images. In order to compare the CapsNet performance in the sub-images, the same classification task was done on different CNN architectures: LeNet (Lecun et al.,1998),AlexNet (Krizhevsky et al.,2012) and VGG-19 (Simonyan& Zisserman,2014). As shown in Table 1, the CapsNet architecture gives a better accuracy. So, for all shuffle testing on the dataset, we obtain an accuracy of 84.7%. We must consider that the main challenge is the dataset, which is noisy and has a low degree of resolution. The main reason why CapsNet gives better results is the fact that the structures of TB cords are floating over the sputum sample and obtaining different 3D shapes. This is where the Caps net performs an essential task, as it is invariant to 3D position changes. In addition, the overall performance of the system based on 20 full images (10 TB positive and 10 TB negative)provides a sensitivity of 80% using the CapsNet architecture for sub-image classification
4. Conclusions
This work shows that the CapsNet architecture provides a classification accuracy of 84.3%, which is better than other CNN architectures such as AlexNet of VGG-19, for TB prediction based on sub image division. Also, this paper demonstrates that our system is capable for prediction ofTB with a sensitivity of 80%, this by using images captured from a lens-free microscopy and using the CapsNet architecture. The results of the test seem promising despite the fact that the images are noisy and low quality. As described in this work, our project contributes to the development of a fast, cost-effective, and globally useful screening tool for tuberculosis detection. The development of such systems helps to improve medical solutions in general, but especially in rural areas where medical resources are limited.
Acknowledgements
This project has been financed by CONCYTEC, by its executing entity FONDECYT, with the objective of promoting the exchange of knowledge between foreign academic institutions and Peru.
References
Arel, I., Rose, D. C., and Karnowski, T. P. Deep machine learning – a new frontier in artificial intelligence research[research frontier]. IEEE Computational Intelligence Magazine, 5(4):13–18, 2010.Iesmantas, T. and Alzbutas, R. Convolutional Capsule Net-work for Classification of Breast Cancer Histology Im-ages, pp. 853–860. 06 2018. ISBN 978-3-319-92999-6.doi: 10.1007/978-3-319-93000-8 97.Krizhevsky, A., Sutskever, I., and Hinton, G. Imagenetclassification with deep convolutional neural networks.Neural Information Processing Systems, 25, 01 2012. doi:10.1145/3065386.Kwabena Patrick, M., Felix Adekoya, A., Abra Mighty, A.,and Edward, B. Y. Capsule networks – a survey. Journalof King Saud University – Computer and InformationSciences, Sep 2019. ISSN 1319-1578.Lecun, Y., Bottou, L., Bengio, Y., and Haffner, P. Gradient-based learning applied to document recognition. Proceed-ings of the IEEE, 86(11):2278–2324, 1998.Mobiny, A., Lu, H., Nguyen, H. V., Roysam, B., andVaradarajan, N. Automated classification of apoptosis inphase contrast microscopy using capsule network. IEEETransactions on Medical Imaging, 39(1):1–10, 2020.Rodr´ıguez, L., Alva, A., Coronel, J., Caviedes, L., Mendoza-Ticona, A., Gilman, R., Sheen, P., and Zimic, M. Imple-mentaci´on de un sistema de telediagn´ostico de tubercu-losis y determinaci´on de multidrogorresistencia basadaen el m´etodo Mods en Trujillo, Per´u. Revista Peruana deMedicina Experimental y Salud Publica, 31:445 – 453,07 2014. ISSN 1726-4634.Sabour, S., Frosst, N., and Hinton, G. E. Dynamic routingbetween capsules, 2017.Simonyan, K. and Zisserman, A. Very deep convolutionalnetworks for large-scale image recognition, 2014.WHO. Global tuberculosis report 2019. In World HealthOrganization report, Geneva, 2019.Zhang, X. and Zhao, S.-G. Fluorescence microscopy imageclassification of 2d hela cells based on the capsnet neuralnetwork. Medical & Biological Engineering & Comput-ing, 57(6):1187–1198, Jun 2019. ISSN 1741-0444. doi:10.1007/s11517-018-01946-z
